PERSONALIZED MEDICINE AND FAST GENOMIC DATA CLEANING
CONTEXT

The last 20 years have seen a drastic drop in the price to sequence a human-sized genome. It was only the early 2000s when the price was about 100 Million US dollars and today, thanks to the advancements of sequencing technologies, the price to obtain digital DNA data of an individual is less than a thousand dollars.
This price drop fostered genomic research to unprecedent development. The possibility to exploit large amount of genomic data in fact, allows the development of today’s personalized medicine with the goal of creating treatments specific on the DNA of each patient, maximizing the efficiency and positive impact on people wellbeing.
Since the price dropped to less than a thousand dollars, the real bottleneck in the genomic data analysis shifted from the acquisition and digitalization of data, to the algorithmic analysis that are today very expensive and time consuming [139^]. To get the final DNA data in fact, it is necessary to go through multiple algorithmic steps that begin with cleaning, correcting and polishing the data provided by the sequencing machine, and terminates with a complete assembly of the genome.
Data Cleaning and Oxford Nanopore Technologies

Among all the algorithmic steps to obtain a full DNA strand, cleaning and correcting reads is of uppermost importance, as it directly impacts the overall quality of the assembled genome. This step impacts more long-read sequencing technologies (that are able to deliver reads longer than 700 base pairs), rather than short ones, given the greater complexity in providing long reads and the errors introduced by the sequencing machines.
In these regards, Oxford Nanopore sequencing technology enables direct, real-time analysis of long DNA or RNA fragments and works by monitoring changes to an electrical current as nucleic acids are passed through a protein nanopore.
The resulting signal is decoded, using software solutions, to provide the specific DNA or RNA sequence, but it can be also accessed to perform custom analysis on the output data.
The advantage of long reads

There are several advantages of using long-read sequencing compared to short-read sequencing. Longer reads allow to identify highly repetitive genomic sequences easier, easing the genome assembly task for large and complex genomes. Long-read sequencing allows the detection of large-scale changes in genomic structure or structural variants, and direct detection of epigenetic modifications, such as DNA methylation, which would require more complex processes using short-read sequencing methods.
There are also few disadvantages in using such technology. The main issue is related to errors in the genomic sequences. These errors are in fact more frequent compared to short-read sequencing technologies and require software algorithms to account for errors and to correct them. Secondly, long-read sequencing technologies provide more data to analyze and require more performant hardware architectures to process the data in a reasonable time, and in the most energy efficient way.
To help software correcting the errors of Oxford Nanopore sequencers, Jared Simpson, a researcher from Ontario Institute for Cancer Research, developed Nanopolish, a software tool that leverages both raw signals coming from the sequencer and basecalled data to help correcting long reads. Huxelerate, enhanced Nanopolish creating Huxelerate Hugenomic Nanopolish, a tool that exploits Xilinx FPGAs to speed up the computational problem, and it solves the computational issue related to analyze the huge amount of data coming from the sequencer.
Nanopolish

Nanopolish is a software package for signal-level analysis of Oxford Nanopore data that exploits the raw signals provided in output by the sequencing machine, to calculate an improved consensus sequence for a draft genome assembly, detect base modifications, call SNPs and indels with respect to a reference genome and more.
In this document, we will focus on a specific module called eventalign, that allows to compare the raw signals provided as output of the sequence machine, to the basecalled data, aligning signal data emitted by a nanopore machine to a reference genome, in contrast to most approaches which align two DNA sequences to each other.
The eventalign tool exploits a Hidden markov Model (HMM) to align signal data from the nanopore sequencing machine to a reference to improve the accuracy of the sequence obtained by the basecaller.
The probabilistic model is used to accomplish the task of calculating the probability of observing an event sequence (a portion of the signal provided in output from the sequencer) given a known DNA sequence (the basecalled sequence).
The HMM used within nanopolish is a profile Hidden Markov Model and helps in calculating the probability of a sequence of events, given a known sequence. Nanopolish handles huge amount of long reads and signals coming from the sequencer.
Huxelerate Hugenomic Nanopolish – ultra fast signal-level analysis

Huxelerate Hugenomic Nanopolish is a software tool built on top of Nanopolish that enables ultra-fast signal-level analysis of large datasets of Oxford Nanopore Sequencing data.
Huxelerate Hugenomic Nanopolish accelerated implementation exploits FPGAs to accelerate the computation and provides high performance and faster time to result of the eventalign module, reducing computational time from days to hours when compared to software-only executions.
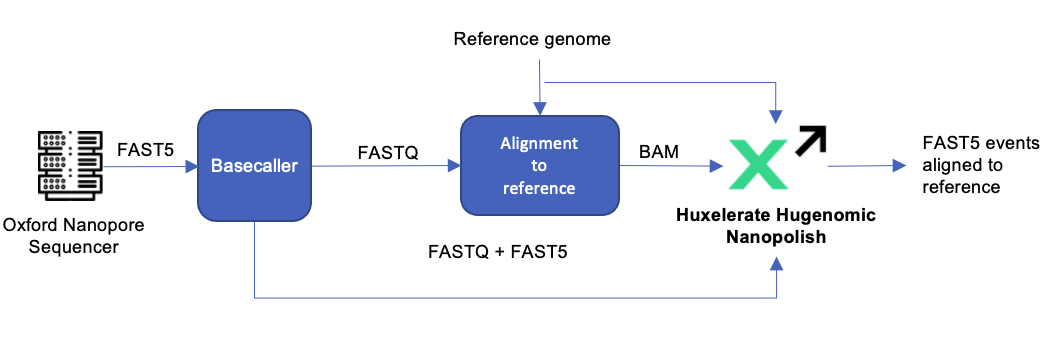
The software is built to be easy to use, and can be used as a replacement of the original Nanopolish, with almost 10 times performance improvement than the original version, when compared to a 36 threads processor.
The inputs for Hugenomic Nanopolish are the FASTQ and FAST5 files from the sequencing machine and the basecaller, the reference genome and the BAM file, and the output provided is the same output provided by the original tool, i.e. the FAST5 events aligned to the reference.
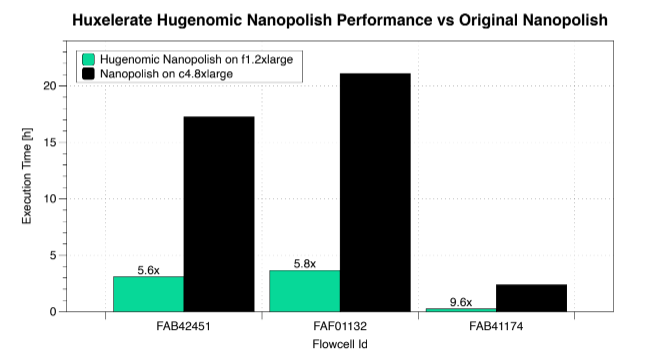
The chart demonstrates the performance improvement when using Huxelerate Hugenomic Nanopolish on an AWS f1.2xlarge instance, equipped with a Xilinx Virtex Ultrascale+ VU9P FPGA compared to a c4.8xlarge compute instance that features a 2.9 GHz Intel Xeon E5-2666 v3 using 36 cores.
The dataset in the experiment came from the WGS Consortium and is available at: https://github.com/nanopore-wgs-consortium/NA12878/blob/master/Genome.md
The accelerated implementation, exploits a hardware architecture on FPGA to accelerate the task of aligning raw signals to basecalled data, exploiting the profile Hidden Markov Model designed for Nanopolish. The CPU and the FPGA work in close collaboration to accomplish the acceleration: the CPU provides data for the FPGA, but it does not idle when waiting for the results, so that an optimal performance value can be reached.
Conclusion

The advent of long-read sequencing technologies enables a whole new set of possible research that were limited by the previous technologies, highlighting the need of more performant software capable of correcting the errors of current technologies, and energy efficient systems to process the ever-increasing amount of data that is coming from the newest sequencing technologies.
To this end, Huxelerate Hugenomic Nanopolish proposes a valid and high performant solution to improve the accuracy of the sequences obtained by Oxford Nanopore basecaller, allowing a more precise and efficient data analysis.
Share on:


